Weight Training Classification
Synopsis
The goal of this project is to correctly classify the quality of physical activity an individual carried out based on accelerometer collected from the belt, forearm, arm, and dumbbell. There were 5 different ways in which the exercise was carried out. It was done either exactly according to the specification (Class A), throwing the elbows to the front (Class B), lifting the dumbbell only halfway (Class C), lowering the dumbbell only halfway (Class D) and throwing the hips to the front (Class E). Class A is the correct manner in which the exercise should be carried out, while the other classes are common errors. Using the accelerometer data I was able to create a random forest model that classified this information into the 5 desired classes with 98 percent accuracy.
library(Hmisc)
library(vip)
library(randomForest)
library(e1071)
library(parallel)
library(MLmetrics)
library(foreach)
library(doParallel)
library(tidytext)
library(tidyverse)
library(reshape2)
library(caret)Read in the Data
#train data location
fileurl<-"https://d396qusza40orc.cloudfront.net/predmachlearn/pml-training.csv"
if (!file.exists("fittrain.csv")){
download.file(fileurl,"fittrain.csv",method="curl")
}
#test data location
fileurl<-"https://d396qusza40orc.cloudfront.net/predmachlearn/pml-testing.csv"
if (!file.exists("fittest.csv")){
download.file(fileurl,"fittest.csv",method="curl")
}
# The data sets had many entries that were spaces ("") and thus I specified these
# to be missing values (NA)
fit_train<-read.csv("fittrain.csv",na.strings = c("NA",""))
fit_test<-read.csv("fittest.csv",na.strings = c("NA",""))Assessing missing data
The first thing I want to do is assess the amount of missing data, so I know if there are certain predictors I should remove, or I should implement an imputation technique.
head(colSums(is.na(fit_train)),n=20)## X user_name raw_timestamp_part_1
## 0 0 0
## raw_timestamp_part_2 cvtd_timestamp new_window
## 0 0 0
## num_window roll_belt pitch_belt
## 0 0 0
## yaw_belt total_accel_belt kurtosis_roll_belt
## 0 0 19216
## kurtosis_picth_belt kurtosis_yaw_belt skewness_roll_belt
## 19216 19216 19216
## skewness_roll_belt.1 skewness_yaw_belt max_roll_belt
## 19216 19216 19216
## max_picth_belt max_yaw_belt
## 19216 19216Given a good number of these predictors are columns of missing values, I will go ahead and remove any predictor that is more than 95% missing values.
Removing Missing Values
threshold=.95
trainfilt<-fit_train[,colSums(is.na(fit_train))<(nrow(fit_train)*threshold)]
testfilt<-fit_test[,colSums(is.na(fit_test))<(nrow(fit_test)*threshold)]Keeping accelerometer data
I am only using data from the accelerometers, as I am interested in assessing whether the acelerometers alone are enough to classify the quality with which an exercise is carried out. As a result, I will remove columns that contain other information.
trainfilt<-trainfilt[,-c(1,2,3,4,5,6,7)]
testfilt<-testfilt[,-c(1,2,3,4,5,6,7)]
trainfilt$classe<-as.factor(trainfilt$classe)
testfilt$classe<-as.factor(testfilt$classe)Near Zero Variance
Next I will examine the data set to determine if there are any uninformative predictors and remove them if that is the case.
nzv<-nearZeroVar(trainfilt,saveMetrics = TRUE)
data.frame(zeroVar=sum(nzv$zeroVar==TRUE), NZV=sum(nzv$nzv==TRUE))## zeroVar NZV
## 1 0 0There are no uninformative predictors, so the data is ready for modelling.
Validation data split
Before modelling, I split the training data set to include a validation set so the model does not overfit.
set.seed(1234)
samp<-createDataPartition(y=trainfilt$classe,p=.8,list = FALSE)
training<-trainfilt[samp,]
validation<-trainfilt[-samp,]Model analysis
The model I am going to use is a random forest model since these typically have superior performance when it comes to classification. I am using a repeated cross validation procedure with 10 folds, and 3 repeats to get a stable model that minimizes overtraining. I will do a grid search to find the optimal value for the mtry parameter. This parameter corresponds to the number of variables randomly sampled as candidates at each split. I center and scale all the predictors and then do a pca selecting the components that account for 95% of the variance. I use a tree size of 250 as this is enough to produce stable accuracy and is not overly computationally intensive. Model performance will be assessed on its accuracy.
Data Preprocessing
control<-trainControl(method='repeatedcv',
number=10,
repeats = 3,
classProbs = TRUE,
summaryFunction = multiClassSummary,
allowParallel = TRUE,
savePredictions = TRUE,
search='grid',
verboseIter = TRUE)
tunevals<-expand.grid(.mtry=c(1:10))
#preprocess all datasets
trainpre<-preProcess(training,method=c('center','scale','pca'),thresh=.95)
trainpca<-predict(trainpre,training)
valpca<-predict(trainpre,validation)
testpca<-predict(trainpre,testfilt)Random Forest Model Fit
#Parallelize the random forest process
cluster<-makeCluster(detectCores()-6)
registerDoParallel(cluster)
set.seed(1234)
rfmod<-train(classe~.,
data=trainpca,
method='rf',
ntree=250,
tuneGrid=tunevals,
verbose=TRUE,
metric='Accuracy',
trControl=control)
stopCluster(cluster)
registerDoSEQ()Optimal mtry
This plot shows how the mtry value affects the cross validation accuracy for the model.
plot(rfmod)
This plot shows that an mtry value of 3 results in the highest accuracy on the held out cross validation sets.
Variable Importance
Using the vip plot we can assess what the most valuable predictors are.
vip(rfmod)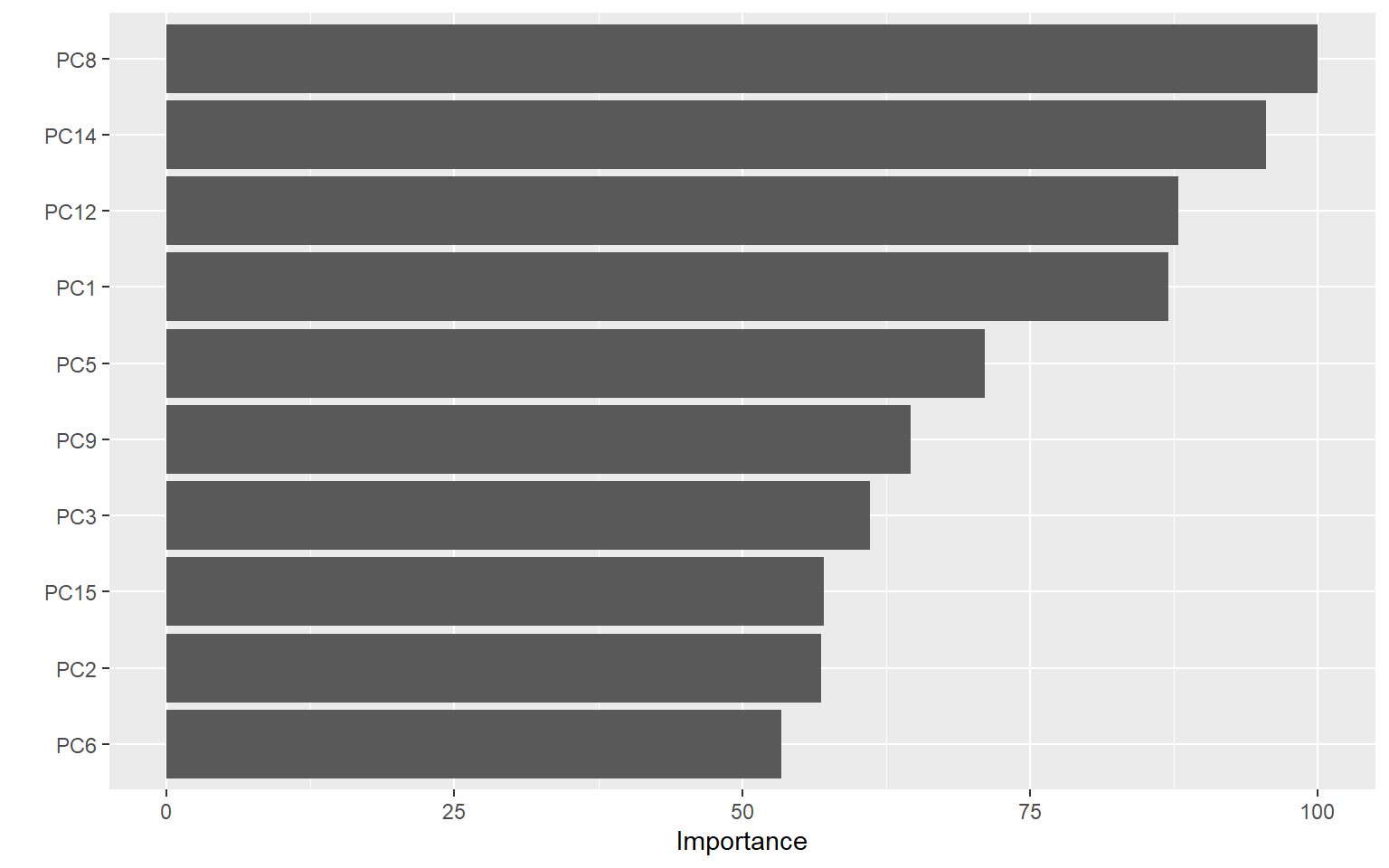
The most important predictor appears to be PC8, so let’s examine that to see what this component is capturing.
Principal Component Analysis
# Extract the pca loadings into an objects that will work with ggplot2
traincomp<-melt(trainpre$rotation)
colnames(traincomp)<-c("Original_Var","Component","Magnitude")
pcaplot<-traincomp %>%
filter(Component %in% paste0("PC", c(8,14,12,1,5))) %>%
ggplot(aes(Magnitude, Original_Var, fill = Original_Var)) +
geom_col(show.legend = FALSE) +
facet_wrap(~Component, nrow = 1) +
labs(y = NULL,
x="Strength of Contribution")
pcaplot
This figure shows the five most informative principal components and the contributions of each predictor into each principal component. Since each component is influenced in some way by each predictor, one way to understand what a component is capturing is by examining the tallest bars in the plot to see what predictors are contributing the most to each component. For example in PC 14 gyroscopic information from the belt in the z and y planes are the largest contributors. Given that one of the classes of exercises involved movement of the hips it makes sense this component would be useful for classification. The following plot shows the most informative predictors that contribute to these components to provide a clearer picture of what these components are capturing.
pcaplot2<-traincomp %>%
filter(Component %in% paste0("PC", c(8,14,12,1,5))) %>%
group_by(Component) %>%
top_n(8, abs(Magnitude)) %>%
ungroup() %>%
mutate(Original_Var = reorder_within(Original_Var, abs(Magnitude), Component)) %>%
ggplot(aes(abs(Magnitude), Original_Var, fill = Magnitude)) +
geom_col() +
scale_fill_viridis_c() +
facet_wrap(~Component, scales = "free_y") +
scale_y_reordered() +
labs(
title="Predictor Contribution to Top Five Principal Components",
x = "Absolute value of contribution",
y = NULL, fill = "Strength of \nContribution"
) +
theme(plot.title=element_text(hjust=.5))
pcaplot2
The strongest predictors that were combined in the 5 most informative principal components are presented in the figure above. Green and yellow colors indicate positive values, while blue and purple colors indicate negative values. The strongest component, PC8, is capturing gyroscopic information from the dumbbell, and the overall acceleration in the forearm arm and forearm in z plane, vs. overall acceleration of the arm, in addition to the magnitude and acceleration of the forearm in the x plane. It makes sense that this is the most important component given it captures so much information about the dumbbell position and the forearm. Both of these are key factors in delineating between correctly and incorrectly carrying out the bicep curl.
ggplot(trainpca,aes(x=PC8,y=PC14,color=classe))+
geom_point(alpha=.3)+
scale_color_brewer(name="Exercise Type",
labels=c("Correct (A)","Elbows Front (B)","Half Curl Lift (C)",
"Half Curl Lower (D)","Hips Front (E)"),
type="qual",palette = "Dark2")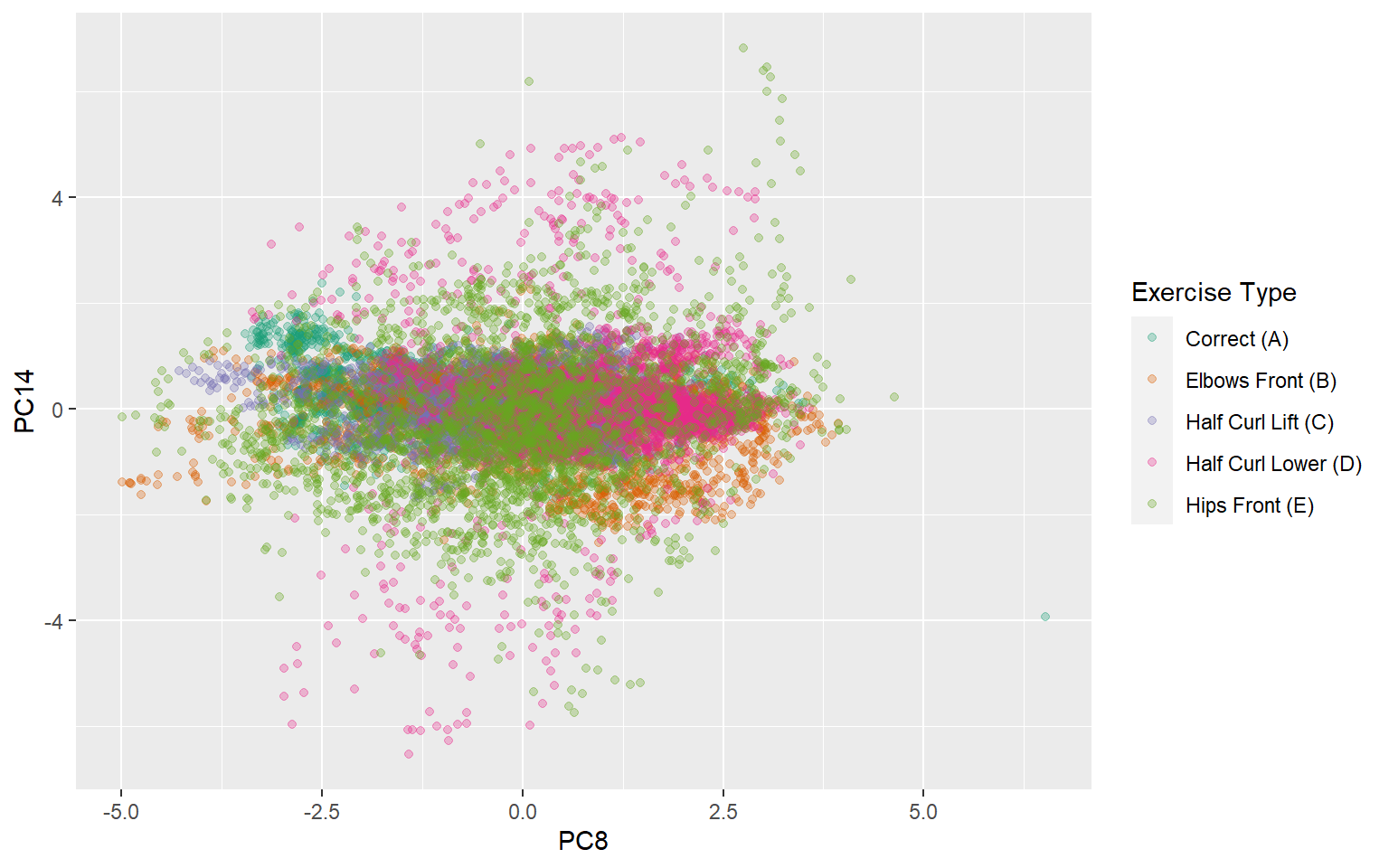
This plot is another way to demonstrate what the two most important principal components are capturing. Since there are so many components and many outcome categories, there are no explicit cutoffs where one component perfectly captures one exercise type. Instead there is a much more complex interaction among the components. PC14 does a relatively good job of capturing some component of exercises where the hips move forward, and the lowering of the dumbbell from halfway. Since PC14 is mainly dominated by gyroscopic information from the belt it makes sense that it would be better at classifying these two types of activities. In the next section I will examine the quality of the model fits.
Model Fit Evaluation
Confusion matrix-caret hold out cross validation data set
To assess the model fit I will be using confusion matrices. First I will examine the model fit to the held out re-samples in the caret cross validation procedure. Then I will examine the model performance against the validation set that I set aside before beginning the modelling.
confusionMatrix(rfmod)## Cross-Validated (10 fold, repeated 3 times) Confusion Matrix
##
## (entries are percentual average cell counts across resamples)
##
## Reference
## Prediction A B C D E
## A 28.2 0.3 0.1 0.0 0.0
## B 0.1 18.8 0.2 0.0 0.1
## C 0.1 0.2 17.0 0.7 0.2
## D 0.0 0.0 0.2 15.6 0.1
## E 0.0 0.0 0.0 0.0 18.0
##
## Accuracy (average) : 0.9767On the cross validation set within the caret procedure, the model is quite accurate (~97%) with few observations off the diagonals.
Confusion matrix-validation data set
confusionMatrix(predict(rfmod,valpca),valpca$classe)## Confusion Matrix and Statistics
##
## Reference
## Prediction A B C D E
## A 1111 10 1 1 0
## B 1 744 12 0 0
## C 2 4 657 25 7
## D 2 0 12 617 6
## E 0 1 2 0 708
##
## Overall Statistics
##
## Accuracy : 0.9781
## 95% CI : (0.973, 0.9824)
## No Information Rate : 0.2845
## P-Value [Acc > NIR] : < 2.2e-16
##
## Kappa : 0.9723
##
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: A Class: B Class: C Class: D Class: E
## Sensitivity 0.9955 0.9802 0.9605 0.9596 0.9820
## Specificity 0.9957 0.9959 0.9883 0.9939 0.9991
## Pos Pred Value 0.9893 0.9828 0.9453 0.9686 0.9958
## Neg Pred Value 0.9982 0.9953 0.9916 0.9921 0.9960
## Prevalence 0.2845 0.1935 0.1744 0.1639 0.1838
## Detection Rate 0.2832 0.1897 0.1675 0.1573 0.1805
## Detection Prevalence 0.2863 0.1930 0.1772 0.1624 0.1812
## Balanced Accuracy 0.9956 0.9881 0.9744 0.9767 0.9905On the validation dataset the model performs fantastic. The model still achieves ~98% accuracy with few observations off the diagonal. The sensitivity of the model is quite high across all classes with .96 being the lowest. Specificity is .99 across all classes. Thus, the model is achieving high accuracy across all classes and is not suffering in its ability to classify one movement over another.
rfpred<-predict(rfmod,testpca)
# Calculate accuracy on the test set
sum(rfpred==testfilt$classe)/nrow(testfilt)*100## [1] 100Conclusion
With the validation set the model performs very well with an accuracy of ~98%. Thus the model has an out of sample error of 2 percent. Given the small size of the test set, the model performs with 100 percent accuracy. Thus this accelerometer data serves as an excellent source of information to classify the quality of an exercise.